Computational Insights into Ludwigia L. Phytochemicals as Potential PDE4B Inhibitors: Molecular Docking and Dynamics Simulations
Main Article Content
Abstract
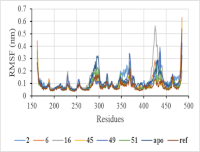
Phosphodiesterase 4B (PDE4B) is an enzyme that regulates inflammatory responses and has recently gained attention as a promising therapeutic target for the treatment of inflammatory diseases. However, the discovery of selective and safe PDE4B inhibitors remains a challenge. The present study aimed to investigate phytochemicals derived from Ludwigia L. as potential PDE4B inhibitors using a combination of computational approaches. A set of compounds was initially screened based on Lipinski’s Rule to assess drug-like properties, with particular emphasis on oral bioavailability and absorption. Toxicity evaluation using LD50 values was performed to categorize the compounds according to safety levels. Molecular docking analysis revealed that several phytochemicals exhibited strong binding affinities toward PDE4B, notably quercetin-3-O-α-L-rhamnoside, luteolin-8-C-glycoside, betulonic acid, (23E)-feruloylhederagenin, and (23Z)-feruloylhederagenin, with compound (23E)-feruloylhederagenin demonstrating the highest docking score (10.88 kcal/mol). Molecular dynamics simulations further confirmed the structural stability of the ligand–protein complexes, with root mean square deviation (RMSD) values consistently below 0.2 nm. Additionally, MMGBSA binding energy calculations supported the strong interaction profile of compound (23E)-feruloylhederagenin, yielding a binding free energy of -54.22 kcal/mol. Taken together, these findings provide computational evidence that Ludwigia L. phytochemicals, particularly (23E)-feruloylhederagenin, represent promising leads for PDE4B inhibition and may serve as valuable candidates for the development of novel anti-inflammatory agents.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1. POWO (2025). Ludwigia L. Plants of the World Online. Facilitated by the Royal BotanicGardens,Kew.Availablefrom: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:30000573-2
2. Kirtikar KR, Basu BD. Indian Medicinal Plants. 2nd ed. India: International Book Distributors; 1999. p. 1088–1089.
3. Ghani A. Medicinal Plants of Bangladesh. 2nd ed. Dhaka: Asiatic Society of Bangladesh; 2003.
4. Perry LM. Medicinal Plants of East and South East Asia: Attributed Properties and Uses. Cambridge, MA: MIT Press; 1980. p. 294.
5. Khan MH, Yadava PS. Antidiabetic plants used in Thoubal district of Manipur, Northeast India. Indian J Tradit Knowl. 2010;9:510–514.
6. Ramírez G, Zavala M, Pérez J, Zamilpa A. In vitro screening of medicinal plants used in Mexico as antidiabetics with glucosidase and lipase inhibitory activities. Evid Based Complement Alternat Med. 2012;2012:701261.
7. Ipor I. Ludwigia octovalvis (Jacq.) P.H. Raven. In: van Valkenburg JLC, Bunyapraphatsara N, editors. Plant Resources of South-East Asia No. 12(2): Medicinal and Poisonous Plants 2. Leiden: Backhuys; 2001.
8. Murugesan T, Sinha S, Pal M, Saha B. Review on phytochemical and medicinal aspects of Jussiaea suberuticosa Linn. Anc Sci Life. 2002;21:205–207.
9. Shaphiullah M, Bachar SC, Kundu JK, Begum F, Uddin MA, Roy SC, Khan M. Antidiarrheal activity of the methanol extract of Ludwigia hyssopifolia Linn. Pak J Pharm Sci. 2003;16(1):7–11.
10. Houslay MD. PDE4 cAMP-specific phosphodiesterases. Cell Signal. 2001;13(9):655–663.
11. Fertig BA, Baillie GS. PDE4-mediated cAMP signalling. J Cardiovasc Dev Dis. 2018;5(1):8.
12. Zervoudakis G, Chou J, Gurney ME, Quesnelle KM. PDE4 subtypes in cancer. Oncogene. 2020;39(19):3791–3802.
13. Menniti FS, Faraci WS, Schmidt CJ. Phosphodiesterases in the CNS: targets for drug development. Nat Rev Drug Discov. 2006;5(8):660–670.
14. Donders Z, Skorupska IJ, Willems E, Mussen F, Van Broeckhoven J, Carlier A. Beyond PDE4 inhibition: A comprehensive review on downstream cAMP signaling in the central nervous system. Biomed Pharmacother. 2024;177:117009.
15. Su Y, Ding J, Yang F, He C, Xu Y, Zhu X. The regulatory role of PDE4B in the progression of inflammatory function study. Front Pharmacol. 2022;13:982130.
16. Blauvelt A, Langley RG, Gordon KB, Silverberg JI, Eyerich K, Sommer MO. Next generation PDE4 inhibitors that selectively target PDE4B/D subtypes: A narrative review. Dermatol Ther. 2023;13(12):3031–3042.
17. Dastidar SG, Rajagopal D, Ray A. Therapeutic benefit of PDE4 inhibitors in inflammatory diseases. Curr Opin Investig Drugs. 2007;8(5):364–372.
18. Fan T, Wang W, Wang Y, Zeng M, Liu Y, Zhu S, Yang L. PDE4 inhibitors: potential protective effects in inflammation and vascular diseases. Front Pharmacol. 2024;15:1407871.
19. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341.
20. Jayaram B, Singh T, Mukherjee G, Mathur A, Shekhar S, Shekhar V. Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics. 2012;13(Suppl 7):S7.
21. Banerjee P, Kemmler E, Dunkel M, Preissner R. ProTox 3.0: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2024;52(W1):W513–W520.
22. Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem. 1999;20(7):720–729.
23. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33.
24. Gewald R, Grunwald C, Egerland U. Discovery of triazines as potent, selective and orally active PDE4 inhibitors. Bioorg Med Chem Lett. 2013;23(15):4308–4314.
25. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461.
26. Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61(8):3891–3898.
27. Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25.
28. Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput. 2021;17(10):6281–6291.
29. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. (Duplicate of ref. 19; nên bỏ 1).
30. Hoang Minh Chau C, Thi Tra Giang N, Thi Thuy Tram N, Thi My Chau L, Xuan Ha N, Thi Thuy P. In silico molecular docking and molecular dynamics of Prinsepia utilis phytochemicals as potential inhibitors of phosphodiesterase 4B. J Chem Res. 2024;48(6):17475198241305879.
31. Gavaldà A, Roberts RS. Phosphodiesterase-4 inhibitors: a review of current developments (2010–2012). Expert Opin Ther Pat. 2013;23(8):997–1016.
32. Kwak HJ, Nam KH. Molecular properties of phosphodiesterase 4 and its inhibition by roflumilast and cilomilast. Molecules. 2025;30(3):692.
33. Filipe HA, Loura LM. Molecular dynamics simulations: advances and applications. Molecules. 2022;27(7):2105.
34. Card GL, England BP, Suzuki Y, Fong D, Powell B, Lee B. Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure. 2004;12(12):2233–2247.
35. Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZ, Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. 2019;119(16):9478–9508.
36. Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461.
37. Shawky EM, Elgindi M, Hassan MM. Phytochemical and biological diversity of genus Ludwigia: A comprehensive review. ERU Res J. 2023;2(3):447–474.
38. Shilpi JA, Gray AI, Seidel V. Chemical constituents from Ludwigia adscendens. Biochem Syst Ecol. 2010;38(1):106–109.
39. Chang CI, Kuo CC, Chang JY, Kuo YH. Three new oleanane-type triterpenes from Ludwigia octovalvis with cytotoxic activity against two human cancer cell lines. J Nat Prod. 2004;67(1):91–93.