Network Pharmacology and Molecular Docking Study of 12-Methoxy-4- Methylvoachalotine (MMV) as an Anticancer Agent
Main Article Content
Abstract
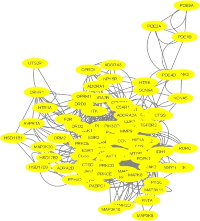
12-Methoxy-4-Methylvoachalotine (MMV) is a sarpagine indole alkaloid isolated from Tabernaemontana macrocarpa and has been implicated in the treatment of tumors and as an anti-snake venom. This study aims to explore MMV as an anticancer agent using in silico methods. The research integrates network pharmacology to identify key protein targets associated with MMV and molecular docking to evaluate the binding affinity and interaction profile of MMV with selected proteins implicated in anticancer mechanisms. The pharmacological network analysis identified ten key targets, including PTEN, HIF1A, CCND1, ESR1, AKT1, MTOR, MCL1, EGFR, and ERBB2, which are closely related to the mechanism of cell proliferation and apoptosis regulation. MMV molecular docking results against the five proteins (TP53, PTEN, ESR1, EGFR, and MCL1) showed lower binding affinity than the native ligands; however, the interaction patterns closely resemble those of the reference ligands. Absorption, distribution, metabolism, excretion, and toxicity (ADMET) predictions indicated low solubility but high gastrointestinal absorption potential. These findings provide important insights into the potential of MMV as an anticancer agent and emphasize the need for structural modifications to enhance its therapeutic efficacy.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1.WHO. Cancer. World Health Organization. 2018; 2:8–9.
2.GCO. Indonesia Cancer Statistics 2022. Global Cancer Observatory, 2022; 149(4): 778–789.
3.Bukowski K, Kciuk M, Kontek R. Mechanisms of multidrug resistance in cancer chemotherapy. Int. J. Mol. Sci. 2020;21(9):3233.: https://doi.org/10.3390/ijms21093233.
4.Amelia P, Nugroho AE, Hirasawa Y, Kaneda T, Tougan T, Horii T, Morita H. Two new sarpagine-type indole alkaloids and antimalarial activity of 16-demethoxycarbonylvoacamine from Tabernaemontana macrocarpa Jack. J. Nat. Med. 2019;73(4):820-5. doi: https://doi.org/10.1007/s11418-019-01317-4.
5.Borges RJ, Cardoso FF, de Carvalho C, de Marino I, Pereira PS, Soares AM, Dal-Pai-Silva M, Usón I, Fontes MR. Structural and functional studies of a snake venom phospholipase A2-like protein complexed to an inhibitor from Tabernaemontana catharinensis. Biochimie. 2023;206:105-15.doi: https://doi.org/10.1016/j.biochi.2022.10.011.
6.Medeiros MR, de Melo Prado LA, Fernandes VC, Figueiredo SS, Coppede J, Martins J, Fiori GM, Martinez-Rossi NM, Beleboni RO, Contini SH, Pereira PS. Antimicrobial activities of indole alkaloids from Tabernaemontana catharinensis. Nat. Prod. Commun. 2011;6(2):1934578X1100600209.
7.Pereira PS, França SD, Oliveira PV, Breves CM, Pereira SI, Sampaio SV, Nomizo A, Dias DA. Chemical constituents from Tabernaemontana catharinensis root bark: a brief NMR review of indole alkaloids and in vitro cytotoxicity. Química Nova. 2008;31:20-4.
8.Bhagat S, Kumar D. A recepto-informatics study of the phytocompounds of Tabernaemontana divaricata (L.) having potential anticancer efficacy for breast cancer. J. Med. Plants Stud. 2021; 9(1). 14–20. doi: https://doi.org/10.22271/plants.2021.v9.i1a.1242.
9.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008 4(11):682-90. doi: https://doi.org/10.1038/nchembio.118.
10.Gunawan PW, Santoso P, Pramitha DAK, Adrianta KA. In Silico Testing of Anti-inflammatory Activity And Toxicity of Peristrophine Compounds On Prostaglandin Synthesis 2 (PTGS2). USADHA: Integrati Obat Tradit. 2025; 1, 1–8, 2025.
11.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L. PubChem 2023 update. Nucleic Acids Res. 2023;51(D1): D1373-80.
12.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357-64.
13.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638-46.
14.Shannon P. Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2023; 13(11): 2498–2504. doi: https://doi.org/10.1101/gr.1239303.
15.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11., doi: https://doi.org/10.1186/1752-0509-8-s4-s11.
16.Hasan R, I’anah FC, Bahi RRR. Molecular Docking of Potential Compounds from Moringa oleifera Leaves Towards Folate Receptors J Innov Res Knowl. 2022; 2(2):519–526. doi: https://doi.org/10.53625/jirk.v2i2.2918.
17.Susanto AF, Jamil AS, Muchlisin MA, Almuhtarihan IF. A network pharmacology of brotowali (Tinospora cordifolia) on immunity cases. In Proceedings of International Pharmacy Ulul Albab Conference and Seminar (PLANAR). 2023;3: 28-36).
18.Susanto AF, Jamil AS, Muchlisin MA, Almuhtarihan IF. A Network Pharmacology of Lemongrass (Cymbopogon citratus) on COVID-19 Cases. In Proceedings of International Pharmacy Ulul Albab Conference and Seminar (PLANAR). 2023;3: 49. doi: https://doi.org/10.18860/planar.v3i0.2471
19.Zhong S, Tian L, Li C, Storch KF, Wong WH. Comparative analysis of gene sets in the gene ontology space under the multiple hypothesis testing framework. PubMed. 2004; 425–35, doi: https://doi.org/10.1109/csb.2004.1332455.
20.Irfandi R, Rijal S, Yani A, Arafah M, Rompegading AB. Literature Review: Network Pharmacology as a New Approach and Trend in Medicine. Hayyan Journal. 2024;1(1):1-8.
21.Li P, Fu Y, Ru J, Huang C, Du J, Zheng C, Chen X, Li P, Lu A, Yang L, Wang Y. Insights from systems pharmacology into cardiovascular drug discovery and therapy. BMC Syst. Biol. 2014;8(1):141. doi: https://doi.org/10.1186/s12918-014-0141-z
22.Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M, Gaudet P, Harris NL, Hill DP. The Gene Ontology Knowledgebase in 2023. Genetics. 2023;224(1):iyad031.
23.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1): D587-92.
24.Rascio F, Spadaccino F, Rocchetti MT, Castellano G, Stallone G, Netti GS, Ranieri E. The pathogenic role of PI3K/AKT pathway in cancer onset and drug resistance: an updated review. Cancers. 2021;13(16):3949.
25.Rah B, Rather RA, Bhat GR, Baba AB, Mushtaq I, Farooq M, Yousuf T, Dar SB, Parveen S, Hassan R, Mohammad F. JAK/STAT signaling: molecular targets, therapeutic opportunities, and limitations of targeted inhibitions in solid malignancies. Front. Pharmacol. 2022;13:821344.
26.Shen J, Wang Q, Mao Y, Gao W, Duan S. Targeting the p53 signaling pathway in cancers: molecular mechanisms and clinical studies. MedComm. 2023;4(3):e288.
27.Yang Y, Ye WL, Zhang RN, He XS, Wang JR, Liu YX, Wang Y, Yang XM, Zhang YJ, Gan WJ. The role of TGF‐β signaling pathways in cancer and its potential as a therapeutic target. J Evid Based Complementary Altern Med. 2021;2021(1):6675208.
28.Park JH, Zhuang J, Li J, Hwang PM. p53 as guardian of the mitochondrial genome. FEBS Lett. 2016;590(7):924-34. doi: https://doi.org/10.1002/1873-3468.12061
29.Khoirunnisa A, Jamil AS, Muchlisin MA. Network Pharmacology Analysis of Secondary Metabolites of Ciplukan (Physalis angulata L.) Against Lung Cancer. Majalah Farmaseutik. 2024;20(2):282-91. doi: https://doi.org/10.22146/farmaseutik.v20i2.96275.
30. Alqarni A, Hosmani J, Alassiri S, Alqahtani AM, Assiri HA. A Network Pharmacology Identified Metastasis Target for Oral Squamous Cell Carcinoma Originating from Breast Cancer with a Potential Inhibitor from F. sargassaceae. Pharmaceuticals. 2024;17(10):1309. doi:https://doi.org/10.3390/ph17101309
31.Okpako IO, Ng’ong’a FA, Kyama CM, Njeru SN. Network pharmacology, molecular docking, and in vitro study on Aspilia pluriseta against prostate cancer. BMC Compl. Med. Therap. 2024;24(1):338. https://doi.org/10.1186/s12906-024-04642-8
32.Rachmania RA. Validation of virtual screening protocol and interaction analysis of natural product-based cancer cell antiproliferation inhibitors against cyclin-dependent kinase 4 (Cdk 4) receptor. Media Farmas. 2019;16(1):21-40.
33.Fitri DA, Hermanto S, Azizah YN. Molecular Docking Study of Bioactive Peptides from Soybean (Glycine max) Resulting from In Silico Hydrolysis of hER-α Receptor (3ERT). Padjadjaran Chemistry. 2023;1(2):122-30.
34.Rastini MB, Giantari NK, Adnyani KD, Laksmiani NP. Molecular docking of anticancer activity of quercetin against breast cancer in silico. J. Chem. 2019;180.doi: https://doi.org/10.24843/jchem.2019.v13.i02.p09
35.Dira Hefnia D, Susantia M, Wahyunia FS, Pratamaa FA. Tetraprenyltoluquinone Inhibits the Tumor Marker Aldo-Keto Reductase: An in Silico Study. Int J Adv Sci Eng Inf Technol. 2022;12(6):2532–2532.doi: https://doi.org/10.18517/ijaseit.12.6.16596.
36.Nainggolan SI, Rajuddin R, Hasanuddin H, Keumalazia R, Hambal M, Frengki F. In silico analysis of anticancer curcumin and its metabolites in increasing the effectiveness of paclitaxel. Res J Pharm Technol. 2023;16(2):885-92. doi: https://doi.org/10.52711/0974-360x.2023.00150