Design, Synthesis, and In Silico Investigation of A Polyheterocyclic Scaffold as A Potential Dual CDK9/Caspase-6 Inhibitor
Main Article Content
Abstract
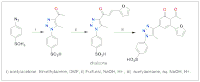
Developing novel therapeutic agents remains a major focus in medicinal chemistry, particularly for cancer and neuroinflammatory disorders. A novel polyheterocyclic compound was synthesised and structurally characterized by FT-IR, ¹H NMR, and ¹³C NMR spectra. To assess its therapeutic potential, an integrated computational approach was conducted, including ligand-based virtual screening, molecular docking, and ADMET predictions. Among five predicted targets, CDK9–Cyclin T1 and Caspase-6 exhibited the most favorable binding profiles, with docking scores of −8.78 kcal/mol and −8.52 kcal/mol, respectively. The compound exhibited key interactions within the active sites of CDK9 and Caspase-6, resembling those of the reference inhibitors Alvocidib and Emricasan. Notably, key interactions with CDK9 involved Glu107 and Asp167, indicating potential for transcriptional inhibition via targets like MYC and MCL-1. Furthermore, the ADME predictions report favourable pharmacokinetic properties, supporting the compound’s candidacy for future biological evaluation as an anticancer and a neuro-antiinflammatory agent.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1.Manwar H, Al-Shuhaib, Z, Hussein KA. Synthesis and molecular docking studies of new tetrazole-acetamide derivatives as anti-cancer agent. Trop J Nat Prod Res. 2024;8(8):8093–8100.
2.Kumar N, Goel N. Heterocyclic compounds: importance in anticancer drug discovery. Anti-Cancer Agents Med. Chem. 2022;22(19):3196–3207.
3.Mateev E, Georgieva M, Zlatkov A. Pyrrole as an important scaffold of anticancer drugs: recent advances. J Pharm Pharm Sci. 2022; (25):24–40.
4.Damayanti D, Iktiarani A, Gymnastiar A. Analysis of active compounds and in silico study of vigna unguiculata as an anti-alzheimer disease agent. Trop J Nat Prod Res. 2025; 9(1):219 -228.
5.Kondeva-Burdina M, Mateev E, Angelov B, Tzankova V, Georgieva M. In silico evaluation and in vitro determination of neuroprotective and MAO-B inhibitory effects of pyrrole-based hydrazones: A therapeutic approach to Parkinson’s disease. Molecules. 2022;27(23):8485.
6.Jeelan N, Basavarajaiah S, Shyamsunder K. Therapeutic potential of pyrrole and pyrrolidine analogs: an update. Mol Divers. 2022;26(5):2915–37. Doi: https://doi.org/10.1007/s11030-022-10387-8.
7.Alghamdi SB, Abdallah WE, Abdelshafeek KA. Assessment of antioxidant, antimicrobial, cytotoxic activities and isolation of some chemical constituents from different extracts of Pergularia daemia. Trop J Nat Prod Res. 2021; 5(10):1816-1827.
8.Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H, Cui Q. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020; 21(6):1960. Doi: https://doi.org/10.3390/ijms21061960.
9.Mandal R, Becker S, Strebhardt K. Targeting CDK9 for anti-cancer therapeutics. Cancers. 2021; 13(9):2181. Doi: https://doi.org/10.3390/cancers13092181.
10.Flores J, Noël A, Fillion ML, LeBlanc AC. Therapeutic potential of Nlrp1 inflammasome, Caspase-1, or Caspase-6 against Alzheimer disease cognitive impairment. Cell Death Differ. 2022; 29(3):657–669.
11.Mazyed H, Nahi R. Synthesis and antioxidant study of new1,3-oxazepin-4,7-dione and1,2,3-triazole derivatives. Int J Pharm Res. 2020; 12(1):252–259.
12.Kozan A, Nahi R. Synthesis And Molecular Docking Studies Of New Pyrimidinone Ring Containing 1,2,3-Triazole Derivatives. Int J Drug Deliv Technol. 2023; 13(3):1005–1010.
13.Kozan A, Nahi R. Combination And Molecular Docking Studies Of 1,2,3-Triazole And Pyrimidin-2-Thione Rings For Potential Anti-COVID-19 Activity. J Heal Soc Sci. 2023; 8(3):249–261.
14.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019; 47(W1):W357–W364. Doi: https://doi.org/10.1093/nar/gkz382.
15.Allouche A. Software news and updates gabedit—a graphical user interface for computational chemistry softwares. J Comput Chem. 2012; 32:174–182. Doi: https://doi.org/10.1002/jcc.21600.
16.Kotynia A, Marciniak A, Kamysz W, Neubauer D, Krzyżak E. Interaction of positively charged oligopeptides with blood plasma proteins. Int J Mol Sci. 2023; 24(3):2836. Doi: https://doi.org/10.3390/ijms24032836.
17.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31(2):455–461. Doi: https://doi.org/10.1002/jcc.21334.
18.Biovia, D. Discovery studio modeling environment, release 2017 Dassault Systèmes. San Diego, CA, USA, 2016.
19.Kartika, I, Syach A, Larasati D, Pitaloka S, Octaviany S, Kurniadewi F, Namirah I. In-silico molecular docking and admet prediction of natural compounds in oncom. Trop J Nat Prod Res. 2024; 8(10942):6737–6740.
20.CCozza G, Zonta F, Vedove A, Venerando A, Dall'Acqua S, Battistutta R, Lolli G. Biochemical and cellular mechanism of protein kinase CK2 inhibition by deceptive curcumin. FEBS J. 2020; 287(9):1850–1864. Doi: https://doi.org/10.1111/febs.15111
21.Freeman B, Hopkins D, Mikochik J, Vacca P, Gao H, Naylor-Olsen A, Trotter W. Discovery of kb-0742, a potent, selective, orally bioavailable small molecule inhibitor of cdk9 for myc-dependent cancers. J Med Chem. 2023; 66(23):15629–15647.
22.Havran M, Chong C, Childers E, Dollings J, Dietrich A, Harrison L, Robichaud J. 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as potent non-peptidic inhibitors of caspase-3. Bioorg Med Chem. 2009; 17(22):7755–7768. Doi: https://doi.org/10.1016/j.bmc.2009.09.036.
23.Wang J, Cao Q, Liu X, Wang T, Mi W, Zhang Y, Su D. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010; 11(11):841–847. Doi: https://doi.org/10.1038/embor.2010.141.
24.Wei Y, Fox T, Chambers P, Sintchak J, Coll T, Golec M, Charifson S. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000; 7(6):423–432.
25.Van S, Wang D, Medina-Cleghorn D, Lee S, Bryant C, Altobelli C, Renslo R. Engaging a non-catalytic cysteine residue drives potent and selective inhibition of caspase-6. J Am Chem Soc. 2023; 145(18):10015–10021.
26.Graham K, Ehrnhoefer E, Hayden R. Caspase-6 and neurodegeneration. Trends Neurosci. 2011;34(12):646–656.
27.Smith E, Soti S, Jones A, Nakagawa A, Xue D, Yin H. Non-steroidal anti-inflammatory drugs are caspase inhibitors. Cell Chem Biol. 2017; 24(3):281–292.
28.Cheng S, Qu Q, Wu, J, Yang J, Liu H, Wang W, Leung H. Inhibition of the CDK9–cyclin T1 protein–protein interaction as a new approach against triple-negative breast cancer. Acta Pharm Sin B. 2022; 12(3):1390–1405. Doi: https://doi.org/10.1016/j.apsb.2021.10.024.
29.Dhani S, Zhao Y, Zhivotovsky B. A long way to go: caspase inhibitors in clinical use. Cell Death Dis. 2021; 12(10):1–13.
30.Mo C, Wei N, Li T, Bhat A, Mohammadi M, Kuang C. CDK9 inhibitors for the treatment of solid tumors. Biochem Pharmacol. 2024; 229:116470. Doi: https://doi.org/10.1016/j.bcp.2024.116470.
31.Tubeleviciute-Aydin A, Beautrait A, Lynham J, Sharma G, Gorelik A, Deny J, LeBlanc C. Identification of Allosteric Inhibitors against Active Caspase-6. Sci Rep. 2019; 9(1):1–19.
32.Eguchi A, Koyama Y, Wree A, Johnson D, Nakamura R, Povero D, Feldstein E. Emricasan, a pan-caspase inhibitor, improves survival and portal hypertension in a murine model of common bile-duct ligation. J Mol Med. 2018; 96(6):575–583.